Update README.md
Browse files
README.md
CHANGED
|
@@ -20,12 +20,12 @@ Custom fine-tuned version of NVIDIA's segformer model for colony slides in micro
|
|
| 20 |
Xie, E., Wang, W., Yu, Z., Anandkumar, A., Alvarez, J. M., & Luo, P. (2021). SegFormer: Simple and efficient design for semantic segmentation with transformers. arXiv preprint arXiv:2105.15203. https://arxiv.org/abs/2105.15203
|
| 21 |
|
| 22 |
The Segformer-Cityscapes model was changed to a ternary classifier and fine-tuned on custom training data.
|
| 23 |
-

|
| 24 |
-

|
| 25 |
-
|
| 26 |
-
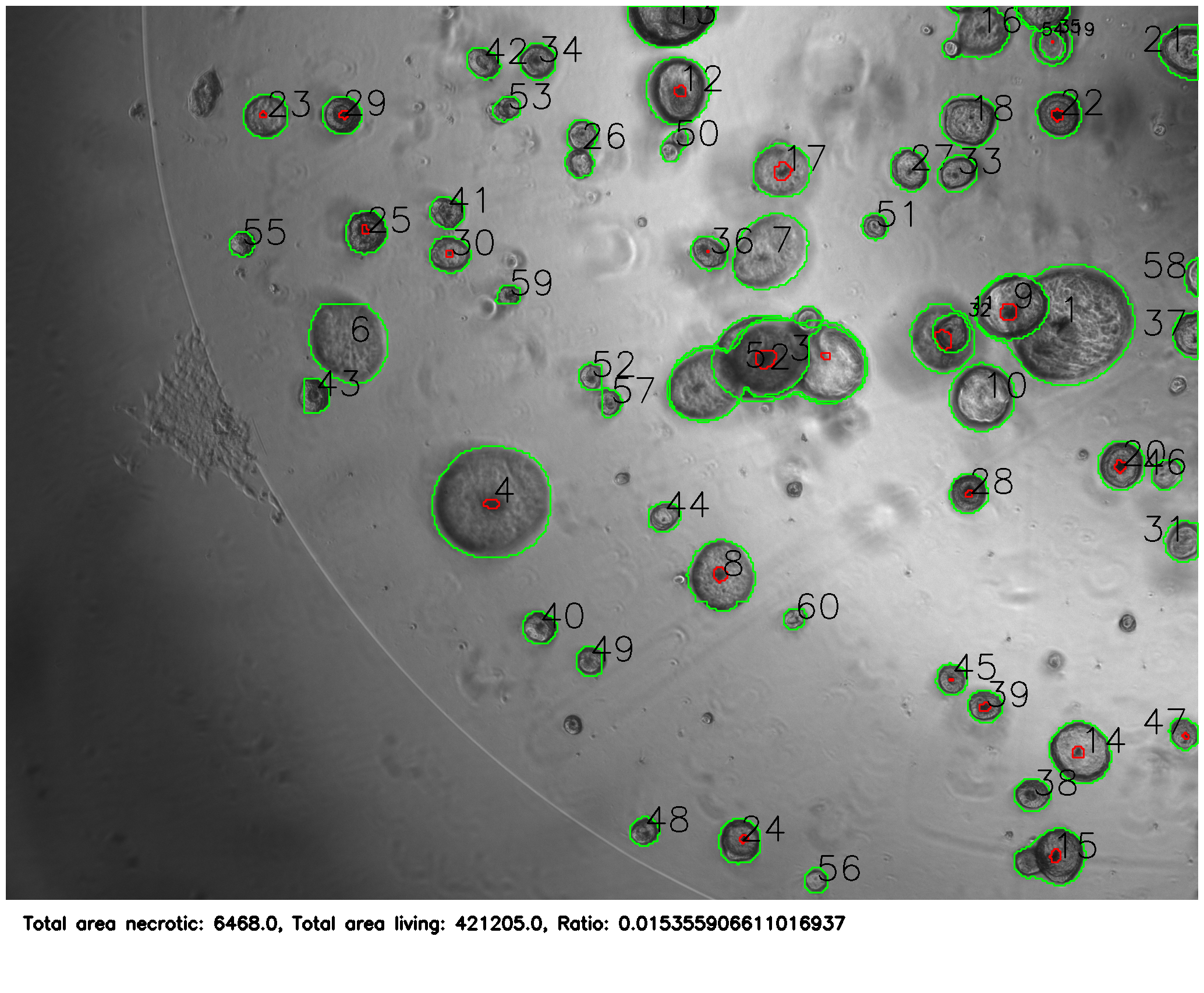
After training, it was able to correctly identify organoids and necrosis.
|
| 27 |

|
| 28 |

|
| 29 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 30 |
The python program (see linked GitHub) then uses the masks to annotate the images and provide statistics about the colonies.
|
| 31 |
|
|
|
|
| 20 |
Xie, E., Wang, W., Yu, Z., Anandkumar, A., Alvarez, J. M., & Luo, P. (2021). SegFormer: Simple and efficient design for semantic segmentation with transformers. arXiv preprint arXiv:2105.15203. https://arxiv.org/abs/2105.15203
|
| 21 |
|
| 22 |
The Segformer-Cityscapes model was changed to a ternary classifier and fine-tuned on custom training data.
|
|
|
|
|
|
|
|
|
|
|
|
|
| 23 |

|
| 24 |

|
| 25 |
|
| 26 |
+
After training, it was able to correctly identify organoids and necrosis.
|
| 27 |
+

|
| 28 |
+

|
| 29 |
+
|
| 30 |
The python program (see linked GitHub) then uses the masks to annotate the images and provide statistics about the colonies.
|
| 31 |

|