File size: 5,334 Bytes
80e841a 14cda03 80e841a de341ba 65371e8 e66c366 65371e8 80e841a d92fafc 80e841a d92fafc 80e841a d92fafc 80e841a e73776a 6f70bd8 015dc2b 388f7f7 80e841a d80de10 ddeb267 80e841a d92fafc 80e841a d92fafc 80e841a d92fafc 80e841a 14cda03 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 |
---
license: mit
datasets:
- chaitjo/QM9_ADiT
- chaitjo/MP20_ADiT
- chaitjo/QMOF150_ADiT
- chaitjo/GEOM-DRUGS_ADiT
tags:
- chemistry
- materials
- molecules
- crystals
- diffusion
- transformer
- latent-diffusion
- all-atom
model-index:
- name: ADiT
results:
- task:
type: unconditional-molecule-generation
dataset:
name: QM9
type: QM9
metrics:
- name: Validity Rate
type: Validity Rate
value: 94.45
source:
name: Unconditional Molecule Generation
url: https://paperswithcode.com/sota/unconditional-molecule-generation-on-qm9
- task:
type: unconditional-crystal-generation
dataset:
name: MP20
type: MP20
metrics:
- name: Validity Rate
type: Validity Rate
value: 91.92
- name: DFT S.U.N. Rate
type: DFT S.U.N. Rate
value: 6
source:
name: Unconditional Crystal Generation
url: https://paperswithcode.com/sota/unconditional-crystal-generation-on-mp20
- task:
type: unconditional-molecule-generation
dataset:
name: GEOM-DRUGS
type: GEOM-DRUGS
metrics:
- name: Validity Rate
type: Validity Rate
value: 95.3
source:
name: Unconditional Molecule Generation
url: https://paperswithcode.com/sota/unconditional-molecule-generation-on-geom
library_name: transformers
---
# All-atom Diffusion Transformers
[](https://www.arxiv.org/abs/2503.03965)
[](https://github.com/facebookresearch/all-atom-diffusion-transformer/)
[](https://huggingface.co/chaitjo/all-atom-diffusion-transformer)
[](https://x.com/chaitjo/status/1899114667219304525)
[](https://www.youtube.com/watch?v=NiY4NLzemnU)
[](https://www.chaitjo.com/publication/joshi-2025-allatom/All_Atom_Diffusion_Transformers_Slides.pdf)
<a target="_blank" href="https://colab.research.google.com/drive/1wHXsP0SHZ-Lx6Brgg-osuvTFrWw3M7oW?usp=sharing">
<img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/>
</a>
Independent reproduction of the paper [*"All-atom Diffusion Transformers: Unified generative modelling of molecules and materials"*](https://www.arxiv.org/abs/2503.03965), by [Chaitanya K. Joshi](https://www.chaitjo.com/), [Xiang Fu](https://xiangfu.co/), [Yi-Lun Liao](https://www.linkedin.com/in/yilunliao), [Vahe Gharakhanyan](https://gvahe.github.io/), [Benjamin Kurt Miller](https://www.mathben.com/), [Anuroop Sriram*](https://anuroopsriram.com/), and [Zachary W. Ulissi*](https://zulissi.github.io/) from FAIR Chemistry at Meta, published at ICML 2025 (* Joint last author).
All-atom Diffusion Transformers (ADiTs) jointly generate both periodic materials and non-periodic molecular systems using a unified latent diffusion framework:
- An autoencoder maps a unified, all-atom representations of molecules and materials to a shared latent embedding space; and
- A diffusion model is trained to generate new latent embeddings that the autoencoder can decode to sample new molecules or materials.
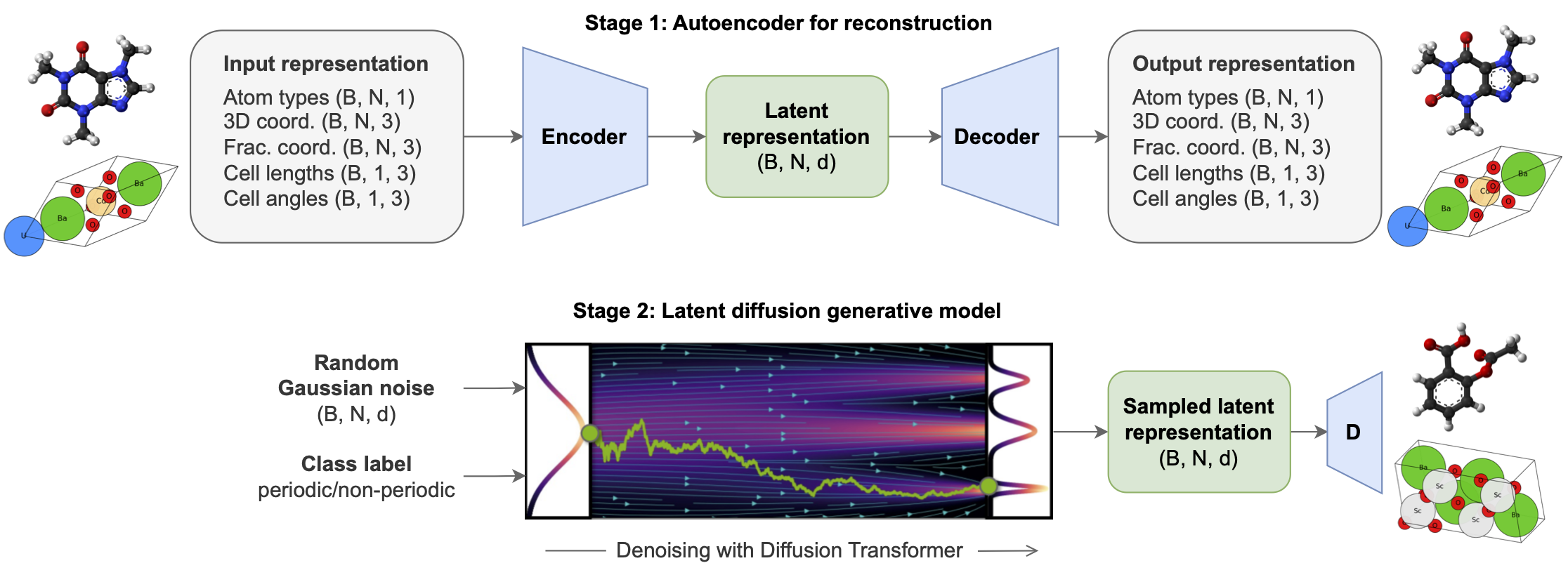
Note that these checkpoints are the result of an independent reproduction of this research by Chaitanya K. Joshi, and may not correspond to the exact models/performance metrics reported in the final manuscript.
These checkpoints can be used to run inference as described in the [README on GitHub](https://github.com/facebookresearch/all-atom-diffusion-transformer/).
Here is a minimal notebook for loading an ADiT checkpoint and sampling some crystals or molecules:
<a target="_blank" href="https://colab.research.google.com/drive/1wHXsP0SHZ-Lx6Brgg-osuvTFrWw3M7oW?usp=sharing">
<img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/>
</a>
Examples of 10,000 sampled crystals and molecules are also available on HuggingFace:
- [Crystals as CIF files](https://huggingface.co/chaitjo/all-atom-diffusion-transformer/resolve/main/ADiT_crystals_mp20.zip) (MP20)
- [Molecules as PDB files](https://huggingface.co/chaitjo/all-atom-diffusion-transformer/resolve/main/ADiT_molecules_qm9.zip) (QM9)
- [Molecules as PDB files](https://huggingface.co/chaitjo/all-atom-diffusion-transformer/resolve/main/ADiT_molecules_geom.zip) (GEOM-DRUGS)
## Citation
Accepted as a conference paper at ICML 2025.
Also presented as a [Spotlight talk](https://www.youtube.com/watch?v=NiY4NLzemnU) at ICLR 2025 AI for Accelerated Materials Design Workshop.
ArXiv link: [*All-atom Diffusion Transformers: Unified generative modelling of molecules and materials*](https://www.arxiv.org/abs/2503.03965)
```
@inproceedings{joshi2025allatom,
title={All-atom Diffusion Transformers: Unified generative modelling of molecules and materials},
author={Chaitanya K. Joshi and Xiang Fu and Yi-Lun Liao and Vahe Gharakhanyan and Benjamin Kurt Miller and Anuroop Sriram and Zachary W. Ulissi},
booktitle={International Conference on Machine Learning},
year={2025},
}
``` |