File size: 1,925 Bytes
86ba0f6 8e20dc8 9989c72 8e20dc8 33ff67c 1263ab8 8e20dc8 4d02925 d7ccd8d a863440 07b90ae cddaf17 1d07707 cb91c3d 1d07707 67be596 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 |
---
license: other
license_name: license.md
license_link: LICENSE
base_model:
- nvidia/segformer-b3-finetuned-cityscapes-1024-1024
tags:
- cancer_research,
- biology,
- microscopy
---
See github here for usage:
https://github.com/ReyaLab/AI_Organoid_Counter
https://reya-lab.org/
We have since updated to [this model](https://huggingface.co/ReyaLabColumbia/Segformer_Organoid_Counter_GP).
Custom fine-tuned version of NVIDIA's segformer model for colony slides in microscopy.
Xie, E., Wang, W., Yu, Z., Anandkumar, A., Alvarez, J. M., & Luo, P. (2021). SegFormer: Simple and efficient design for semantic segmentation with transformers. arXiv preprint arXiv:2105.15203. https://arxiv.org/abs/2105.15203
This model takes 512x512 images for segmentation.
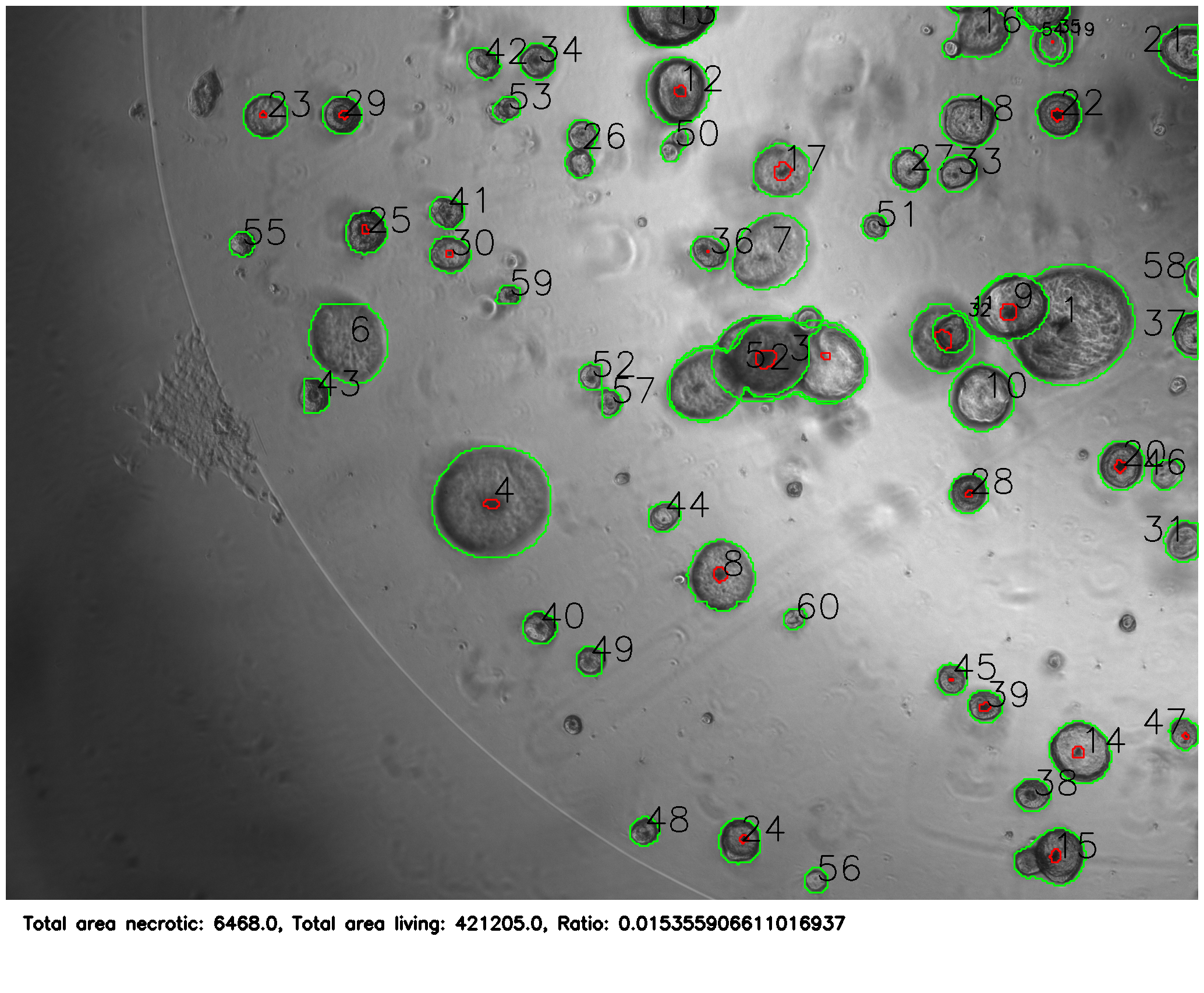
The Segformer-Cityscapes model was changed to a ternary classifier and fine-tuned on custom training data, where organoids and necrosis were made as separate masks and then merged with different grayscale values.



After training, it was able to correctly identify organoids and necrosis.


The python program (see linked GitHub) then uses the masks to annotate the images and provide statistics about the colonies.
 |