LLaVA-Rad
This is the official model checkpoint for LLaVA-Rad, described in “A Clinically Accessible Small Multimodal Radiology Model and Evaluation Metric for Chest X-Ray Findings”.
| Developed by | Microsoft Research |
| Model type | small multimodal transformer model |
| Languages | English |
| License |  |
| Data |  |
| Code |  |
| Evaluation |  |
| Preprint |  |
| Peer Reviewed Paper |  |
LlaVA-Rad is a 7 billion parameter small multimodal model trained to produce findings given an input chest X-ray. Its architecture follows that of LLaVA and LLaVA-Med, differing in the use of a specialized chest X-ray image encoder, BiomedCLIP-CXR, built with the BiomedCLIP framework. LLaVA-Rad offers outstanding performance at relatively small model size.
Contents
Usage
Installation
Follow these steps to set up LLaVA-Rad:
- Clone the repository and navigate to the project folder:
git clone https://github.com/microsoft/llava-rad.git cd llava-rad - Create and activate a virtual environment (Python 3.10):
conda create -n llavarad python=3.10 -y conda activate llavarad - Upgrade pip and install the package:
pip install --upgrade pip pip install -e .
Example inference
import requests
import torch
from PIL import Image
from io import BytesIO
from llava.constants import IMAGE_TOKEN_INDEX
from llava.conversation import conv_templates
from llava.model.builder import load_pretrained_model
from llava.utils import disable_torch_init
from llava.mm_utils import tokenizer_image_token, KeywordsStoppingCriteria
def load_image(image_file):
if image_file.startswith('http') or image_file.startswith('https'):
response = requests.get(image_file)
image = Image.open(BytesIO(response.content)).convert('RGB')
else:
image = Image.open(image_file).convert('RGB')
return image
# Model
disable_torch_init()
model_path = "microsoft/llava-rad"
model_base = "lmsys/vicuna-7b-v1.5"
model_name = "llavarad"
conv_mode = "v1"
tokenizer, model, image_processor, context_len = load_pretrained_model(model_path, model_base, model_name)
# Prepare query
image_file = "https://openi.nlm.nih.gov/imgs/512/253/253/CXR253_IM-1045-1001.png" # CXR w pneumothorax from Open-I
query = "<image>\nDescribe the findings of the chest x-ray.\n"
conv = conv_templates[conv_mode].copy()
conv.append_message(conv.roles[0], query)
conv.append_message(conv.roles[1], None)
prompt = conv.get_prompt()
image = load_image(image_file)
image_tensor = image_processor.preprocess(image, return_tensors='pt')['pixel_values'][0].half().unsqueeze(0).cuda()
input_ids = tokenizer_image_token(prompt, tokenizer, IMAGE_TOKEN_INDEX, return_tensors='pt').unsqueeze(0).cuda()
stopping_criteria = KeywordsStoppingCriteria(["</s>"], tokenizer, input_ids)
with torch.inference_mode():
output_ids = model.generate(
input_ids,
images=image_tensor,
do_sample=False,
temperature=0.0,
max_new_tokens=1024,
use_cache=True,
stopping_criteria=[stopping_criteria])
outputs = tokenizer.batch_decode(output_ids[:, input_ids.shape[1]:], skip_special_tokens=True)[0]
outputs = outputs.strip()
print(outputs)
# Large left pneumothorax is present with apical pneumothorax component
# measuring approximately 3.4 cm in craniocaudal dimension, and a basilar
# component overlying the left hemidiaphragm, with visceral pleural line just
# below the left seventh posterior rib. Cardiomediastinal contours are normal.
# The lungs are clear. No pleural effusion.
Usage and License Notices
The model’s intended use is to generate draft findings of chest X-ray images in English. It is provided for reproducibility and to enable further research.
The data, code, and model checkpoints are licensed and intended for research use only. The code and model checkpoints are subject to additional restrictions as determined by the Terms of Use of LLaMA, Vicuna, and GPT-4 respectively. Code and model checkpoints may be used for research purposes only and should not be used in direct clinical care or for any clinical decision making purpose. You bear sole responsibility for use of the code and model checkpoints, including incorporation into any product used for clinical purposes.
Ethical Considerations and Limitations
Microsoft believes Responsible AI is a shared responsibility and we have identified six principles and practices to help organizations address risks, innovate, and create value: fairness, reliability and safety, privacy and security, inclusiveness, transparency, and accountability. When downloaded or used in accordance with our terms of service, developers should work with their supporting model team to ensure this model meets requirements for the relevant use case and addresses unforeseen product misuse. While testing the model with images and/or text, ensure that the data is PHI free and that there are no patient information or information that can be tracked to a patient identity.
Out-of-scope use
The model is NOT designed for the following use cases:
* Use by clinicians to inform clinical decision-making, as a diagnostic tool or as a medical device. Although LLaVA-Rad attains state-of-the-art performance, it is not designed or intended to be deployed in clinical settings as-is nor is it for use in the diagnosis, cure, mitigation, treatment, or prevention of disease or other conditions (including to support clinical decision-making), or as a substitute of professional medical advice, diagnosis, treatment, or clinical judgment of a healthcare professional.
* Scenarios without consent for data - Any scenario that uses health data for a purpose for which consent was not obtained.
* Use outside of health scenarios - Any scenario that uses non-medical related image and/or serving purposes outside of the healthcare domain.
Please see Microsoft's Responsible AI Principles and approach available at https://www.microsoft.com/en-us/ai/principles-and-approach/
Data provenance and potential biases
BiomedCLIP-CXR was trained with datasets containing chest X-ray images acquired from adult patients in the United States (Stanford Hospital in CA, Beth Israel Deaconess Medical Center in MA), New Zealand, Brazil, Vietnam, Spain, and China. Synthetic free-text reports were generated for datasets without readily available corresponding reports, or translated from other languages such as Spanish into English.
The projector and decoder layers of LLaVA-Rad were trained using MIMIC-CXR data, containing English only reports for patients from Boston, MA. Please see our manuscript for references to individual publications that describe demographic characteristics of patients from whom chest X-ray images were obtained in detail.
Radiology reporting styles can vary within and across health systems and regions. This may impact the generalizability of the model to unseen populations and as such the model should always be tested on the intended deployment population.
This model may produce errors in the generated findings (see CheXprompt evaluation and other results in manuscript for details).
LLaVA-Rad Basics
Abstract
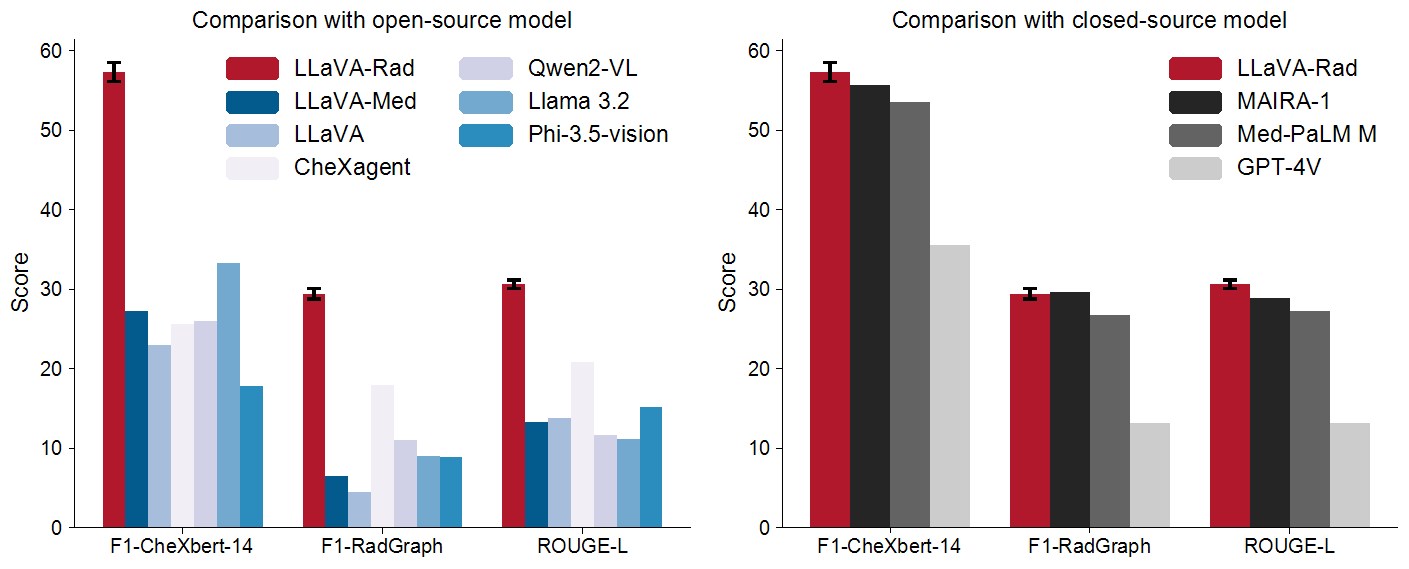
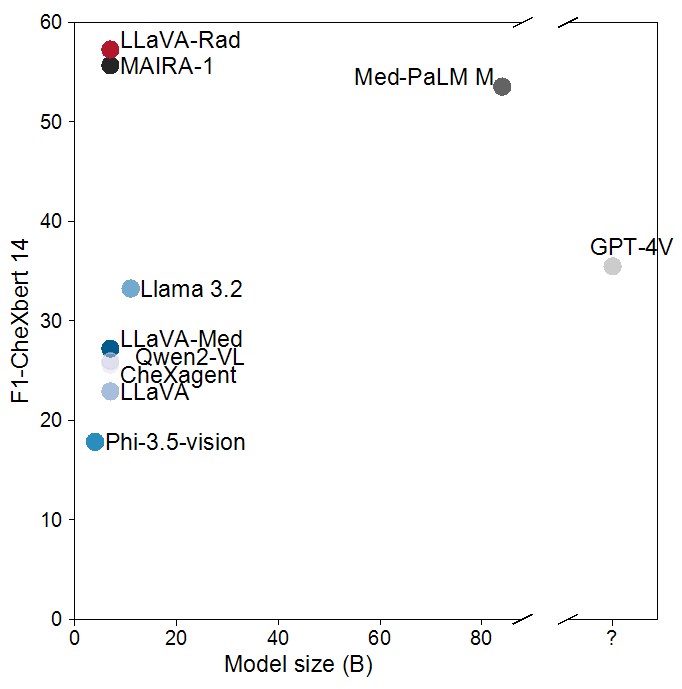
The scaling laws and extraordinary performance of large foundation models motivate the development and utilization of such models in biomedicine. However, despite early promising results on some biomedical benchmarks, there are still major challenges that need to be addressed before these models can be used in real-world clinics. This work aims to address these challenges by training small multimodal models (SMMs) to bridge competency gaps for unmet clinical need in radiology. To maximize data efficiency, we adopt a modular approach by incorporating state-of-the-art pre-trained models for image and text modalities, and focusing on training a lightweight adapter to ground each modality to the text embedding space. For training, we assemble a large dataset of over 697 thousand radiology image-text pairs. LLaVA-Rad (7B) model attains state-of-the-art results on standard radiology tasks such as report generation and cross-modal retrieval, even outperforming much larger models such as GPT-4V and Med-PaLM M (84B). Moreover, LLaVA-Rad requires only one day to be trained on over 697 thousand image-text pairs using a standard 8-A100 GPU cluster, allowing further fine-tuning by clinicians using their own data. The inference of LLaVA-Rad is fast and can be performed on a single V100 GPU in private settings, offering a promising state-of-the-art tool for real-world clinical applications.
Model Architecture
LLaVA-Rad follows the LLaVA v1.5 architecture, which employs an image encoder, a transformer-based language decoder, and a multilayer perceptron connector. For the image encoder, it uses a custom model named BiomedCLIP-CXR, based on BiomedCLIP. For the language decoder it uses lmsys/vicuna-7b-v1.5.
Evaluation Results
To reproduce the results above, please follow the steps outlined in the official code implementation: setup, inference and eval .
Please see the paper for detailed information about methods and results: https://arxiv.org/pdf/2412.10337.
You may also find the CheXprompt library useful for evaluation of generated reports. It is accessible at https://github.com/microsoft/chexprompt.
To use this model to reproduce results in the manuscript, follow these steps using the supported code:
Preparation Before running the commands below, you need to have the data, image folder, and the above checkpoints ready.
- Text Data
To download the data, sign the data use agreement and follow the instructions for download at LLaVA-Rad MIMIC-CXR Annotations on PhysioNet. This will include reports with extracted sections in LLaVA format, split into train/dev/test.
- Images
You need to download the MIMIC-CXR-JPG images from PhysioNet by signing the data use agreement and following the instructions.
- Model weights You can find the pretrained model weights for BiomedCLIP-CXR and LLaVA-Rad at https://huggingface.co/microsoft/llava-rad (this repository).
Before proceeding, change the paths in the scripts below according to where you downloaded the data. Batch size is set for 4-GPU machines. If your machine has a difference number of GPUs, please change batch size.
- Inference
To perform inference, update the paths in the evaluation script, found in the official codebase.
bash scripts/eval.sh
- Evaluation
If you have run inference using multiple GPUs and have a resulting set of chunks with results, make sure you concatenate prediction chunks into a single file before running the following command:
cd llava/eval/rr_eval
python run.py ${YOUR_PREDICTION_FILE}
Citation
Please cite our paper if you use the code, model, or data.
Zambrano Chaves JM, Huang S-C, Xu Y, Xu H, Usuyama N, Zhang S, et al. Towards a clinically accessible radiology foundation model: open-access and lightweight, with automated evaluation. arXiv preprint arXiv:2403.08002. 2024.
Bibtex:
@article{zambranochaves2024llavarad,
title = {Towards a clinically accessible radiology foundation model: open-access and lightweight, with automated evaluation},
author = {Zambrano Chaves, JM and Huang, S-C and Xu, Y and Xu, H and Usuyama, N and Zhang, S, et al.},
journal = {arXiv preprint arXiv:2403.08002},
year = {2024},
url = {https://arxiv.org/pdf/2403.08002}
}
Further Information
Data Specification
Additional details regarding the data used can be found in the corresponding data release, LLaVA-Rad MIMIC-CXR Annotations: https://physionet.org/content/llava-rad-mimic-cxr-annotation/1.0.0/
Related Models
CxrReportGen (https://aka.ms/CXRReportGenModelCard) offers similar functionality based on the MAIRA-2 framework.
- Downloads last month
- 2,650