|
|
--- |
|
|
license: mit |
|
|
datasets: |
|
|
- chaitjo/QM9_ADiT |
|
|
- chaitjo/MP20_ADiT |
|
|
- chaitjo/QMOF150_ADiT |
|
|
- chaitjo/GEOM-DRUGS_ADiT |
|
|
tags: |
|
|
- chemistry |
|
|
- materials |
|
|
- molecules |
|
|
- crystals |
|
|
- diffusion |
|
|
- transformer |
|
|
- latent-diffusion |
|
|
- all-atom |
|
|
model-index: |
|
|
- name: ADiT |
|
|
results: |
|
|
- task: |
|
|
type: unconditional-molecule-generation |
|
|
dataset: |
|
|
name: QM9 |
|
|
type: QM9 |
|
|
metrics: |
|
|
- name: Validity Rate |
|
|
type: Validity Rate |
|
|
value: 94.45 |
|
|
source: |
|
|
name: Unconditional Molecule Generation |
|
|
url: https://paperswithcode.com/sota/unconditional-molecule-generation-on-qm9 |
|
|
- task: |
|
|
type: unconditional-crystal-generation |
|
|
dataset: |
|
|
name: MP20 |
|
|
type: MP20 |
|
|
metrics: |
|
|
- name: Validity Rate |
|
|
type: Validity Rate |
|
|
value: 91.92 |
|
|
- name: DFT S.U.N. Rate |
|
|
type: DFT S.U.N. Rate |
|
|
value: 6 |
|
|
source: |
|
|
name: Unconditional Crystal Generation |
|
|
url: https://paperswithcode.com/sota/unconditional-crystal-generation-on-mp20 |
|
|
- task: |
|
|
type: unconditional-molecule-generation |
|
|
dataset: |
|
|
name: GEOM-DRUGS |
|
|
type: GEOM-DRUGS |
|
|
metrics: |
|
|
- name: Validity Rate |
|
|
type: Validity Rate |
|
|
value: 95.3 |
|
|
source: |
|
|
name: Unconditional Molecule Generation |
|
|
url: https://paperswithcode.com/sota/unconditional-molecule-generation-on-geom |
|
|
library_name: transformers |
|
|
--- |
|
|
|
|
|
# All-atom Diffusion Transformers |
|
|
|
|
|
[](https://www.arxiv.org/abs/2503.03965) |
|
|
[](https://github.com/facebookresearch/all-atom-diffusion-transformer/) |
|
|
[](https://huggingface.co/chaitjo/all-atom-diffusion-transformer) |
|
|
[](https://x.com/chaitjo/status/1899114667219304525) |
|
|
[](https://www.youtube.com/watch?v=NiY4NLzemnU) |
|
|
[](https://www.chaitjo.com/publication/joshi-2025-allatom/All_Atom_Diffusion_Transformers_Slides.pdf) |
|
|
<a target="_blank" href="https://colab.research.google.com/drive/1wHXsP0SHZ-Lx6Brgg-osuvTFrWw3M7oW?usp=sharing"> |
|
|
<img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/> |
|
|
</a> |
|
|
|
|
|
|
|
|
Independent reproduction of the paper [*"All-atom Diffusion Transformers: Unified generative modelling of molecules and materials"*](https://www.arxiv.org/abs/2503.03965), by [Chaitanya K. Joshi](https://www.chaitjo.com/), [Xiang Fu](https://xiangfu.co/), [Yi-Lun Liao](https://www.linkedin.com/in/yilunliao), [Vahe Gharakhanyan](https://gvahe.github.io/), [Benjamin Kurt Miller](https://www.mathben.com/), [Anuroop Sriram*](https://anuroopsriram.com/), and [Zachary W. Ulissi*](https://zulissi.github.io/) from FAIR Chemistry at Meta, published at ICML 2025 (* Joint last author). |
|
|
|
|
|
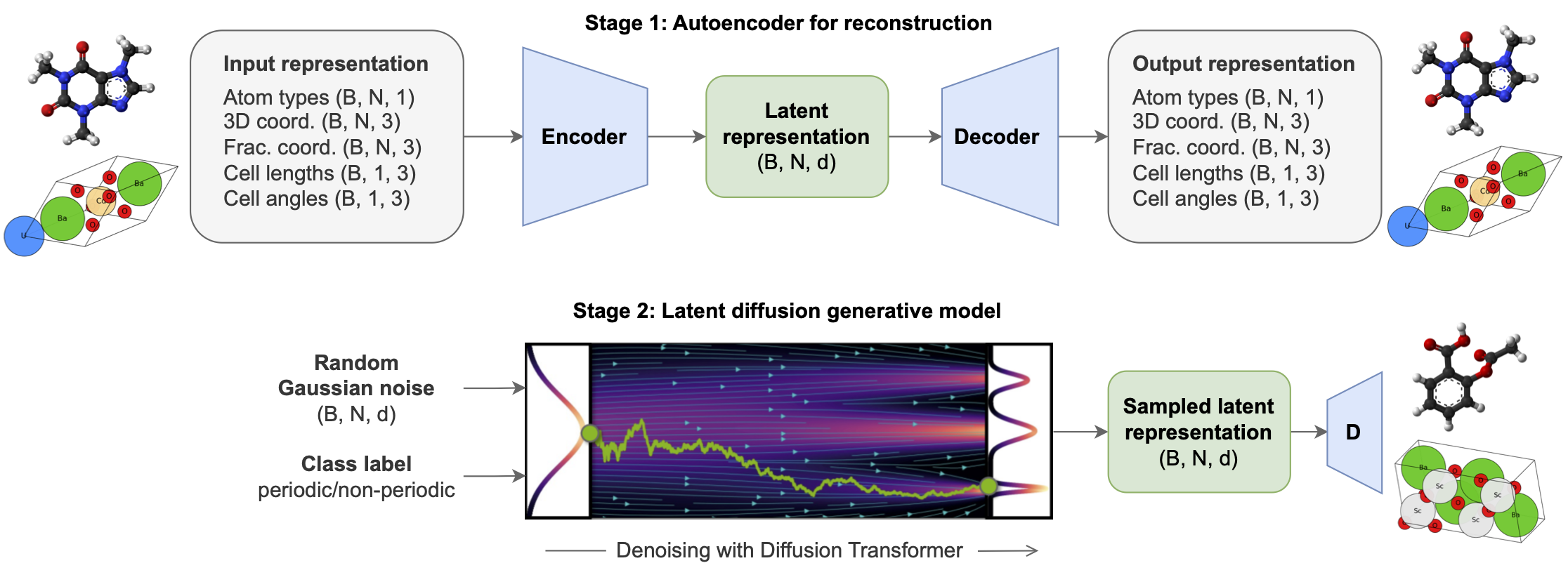
All-atom Diffusion Transformers (ADiTs) jointly generate both periodic materials and non-periodic molecular systems using a unified latent diffusion framework: |
|
|
- An autoencoder maps a unified, all-atom representations of molecules and materials to a shared latent embedding space; and |
|
|
- A diffusion model is trained to generate new latent embeddings that the autoencoder can decode to sample new molecules or materials. |
|
|
|
|
|
 |
|
|
|
|
|
Note that these checkpoints are the result of an independent reproduction of this research by Chaitanya K. Joshi, and may not correspond to the exact models/performance metrics reported in the final manuscript. |
|
|
|
|
|
These checkpoints can be used to run inference as described in the [README on GitHub](https://github.com/facebookresearch/all-atom-diffusion-transformer/). |
|
|
|
|
|
Here is a minimal notebook for loading an ADiT checkpoint and sampling some crystals or molecules: |
|
|
<a target="_blank" href="https://colab.research.google.com/drive/1wHXsP0SHZ-Lx6Brgg-osuvTFrWw3M7oW?usp=sharing"> |
|
|
<img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/> |
|
|
</a> |
|
|
|
|
|
Examples of 10,000 sampled crystals and molecules are also available on HuggingFace: |
|
|
- [Crystals as CIF files](https://huggingface.co/chaitjo/all-atom-diffusion-transformer/resolve/main/ADiT_crystals_mp20.zip) (MP20) |
|
|
- [Molecules as PDB files](https://huggingface.co/chaitjo/all-atom-diffusion-transformer/resolve/main/ADiT_molecules_qm9.zip) (QM9) |
|
|
- [Molecules as PDB files](https://huggingface.co/chaitjo/all-atom-diffusion-transformer/resolve/main/ADiT_molecules_geom.zip) (GEOM-DRUGS) |
|
|
|
|
|
## Citation |
|
|
|
|
|
Accepted as a conference paper at ICML 2025. |
|
|
Also presented as a [Spotlight talk](https://www.youtube.com/watch?v=NiY4NLzemnU) at ICLR 2025 AI for Accelerated Materials Design Workshop. |
|
|
ArXiv link: [*All-atom Diffusion Transformers: Unified generative modelling of molecules and materials*](https://www.arxiv.org/abs/2503.03965) |
|
|
|
|
|
``` |
|
|
@inproceedings{joshi2025allatom, |
|
|
title={All-atom Diffusion Transformers: Unified generative modelling of molecules and materials}, |
|
|
author={Chaitanya K. Joshi and Xiang Fu and Yi-Lun Liao and Vahe Gharakhanyan and Benjamin Kurt Miller and Anuroop Sriram and Zachary W. Ulissi}, |
|
|
booktitle={International Conference on Machine Learning}, |
|
|
year={2025}, |
|
|
} |
|
|
``` |