metadata
license: mit
tags:
- biology
size_categories:
- 10M<n<100M
configs:
- config_name: expression_data
data_files: data/train-*
- config_name: sample_metadata
data_files: metadata/sample_metadata.parquet
- config_name: gene_vocabulary
data_files: metadata/gene_vocabulary.json
- config_name: drug_metadata
data_files: metadata/drug_metadata.parquet
- config_name: cell_line_metadata
data_files: metadata/cell_line_metadata.parquet
dataset_info:
features:
- name: genes
sequence: int64
- name: expressions
sequence: float32
- name: drug
dtype: string
- name: sample
dtype: string
- name: BARCODE_SUB_LIB_ID
dtype: string
- name: cell_line_id
dtype: string
- name: moa-fine
dtype: string
- name: canonical_smiles
dtype: string
- name: pubchem_cid
dtype: string
- name: plate
dtype: string
splits:
- name: train
num_bytes: 1693653078843
num_examples: 95624334
download_size: 337644770670
dataset_size: 1693653078843
Tahoe-100M
Tahoe-100M is a giga-scale single-cell perturbation atlas consisting of over 100 million transcriptomic profiles from 50 cancer cell lines exposed to 1,100 small-molecule perturbations. Generated using Vevo Therapeutics' Mosaic high-throughput platform, Tahoe-100M enables deep, context-aware exploration of gene function, cellular states, and drug responses at unprecedented scale and resolution. This dataset is designed to power the development of next-generation AI models of cell biology, offering broad applications across systems biology, drug discovery, and precision medicine.
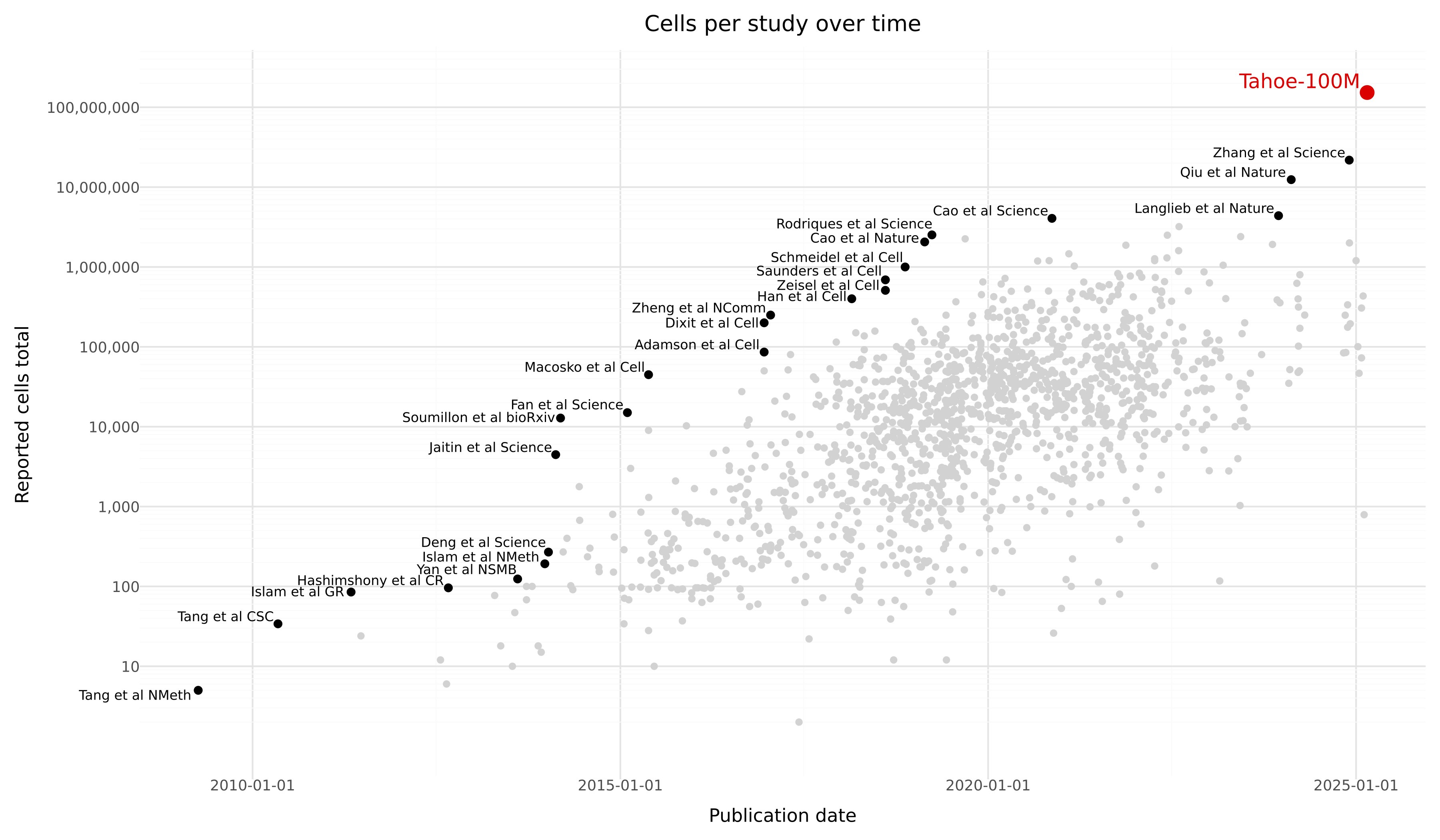
Dataset Description
Features
The dataset has the following fields:
Additional metadata
How to use
Citation
Please cite:
@article{zhang2025tahoe,
title={Tahoe-100M: A Giga-Scale Single-Cell Perturbation Atlas for Context-Dependent Gene Function and Cellular Modeling},
author={Zhang, Jesse and Ubas, Airol A and de Borja, Richard and Svensson, Valentine and Thomas, Nicole and Thakar, Neha and Lai, Ian and Winters, Aidan and Khan, Umair and Jones, Matthew G and others},
journal={bioRxiv},
pages={2025--02},
year={2025},
publisher={Cold Spring Harbor Laboratory}
}